How SNAPc recruits human RNA polymerases at snRNA genes
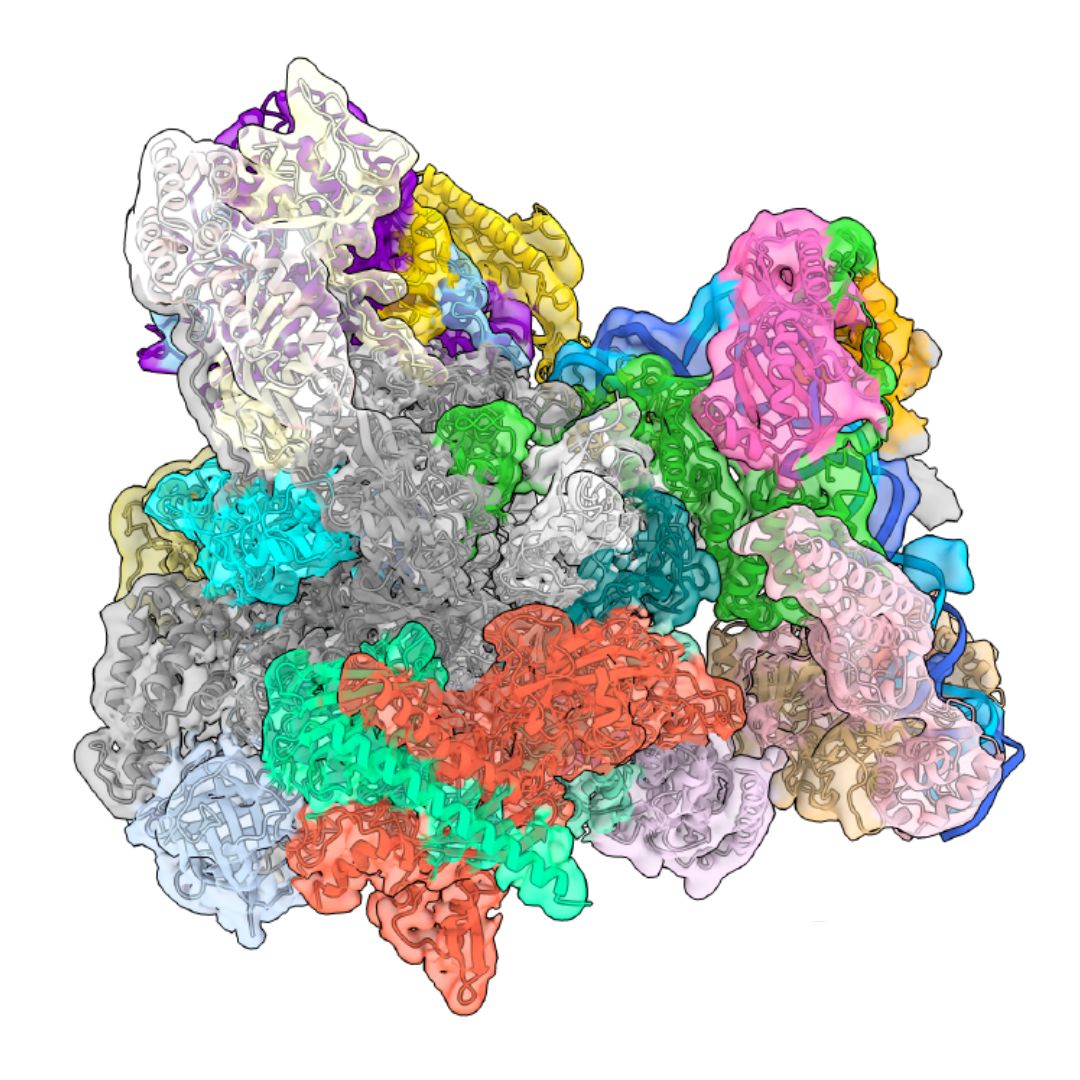
Using advanced cryo-electron microscopy and cross-linking mass spectrometry, HT researchers provide unprecedented insight into how SNAPc works as a “dual recruiter” for RNA polymerase II and III. The findings are published in Nature Communications.
RNA polymerases are tiny molecular machines that produce RNAs, essential molecules for life. Among these, RNA polymerase III (Pol III) transcribes small but crucial RNAs such as U6 snRNA, which plays a key role in RNA splicing – a process critical for producing functional proteins. Pol III dysregulation leads to several diseases, including Alzheimer’s disease and cancer.
The recruitment of Pol III to specific U6 snRNA genes requires a molecular complex called SNAPc. In addition, SNAPc can recruit RNA polymerase II (Pol II), thus mediating the synthesis of different types of snRNA. How SNAPc specifically binds either polymerase while maintaining a precise function remains poorly understood.
Alessandro Vannini and Ewan Ramsay – Head of the Structural Biology Research Centre and senior staff scientist at Human Technopole, respectively – exploited cutting-edge cryo-electron microscopy (cryo-EM) to visualise the structure of SNAPc in complex with the U6 snRNA promoter. The work was performed in collaboration with researchers at the Institute of Cancer Research, London (UK) and the Structural and Computational Biology Unit at EMBL (Germany).
The cryo-EM structure of SNAPc unveiled that an adaptable structural design drives SNAPc’s ability to recruit different RNA polymerases. SNAPc can flip its orientation depending on the type of recruited RNA polymerase, thus ensuring the correct polymerase is engaged at the right site. The structure also revealed that SNAPc subunit SNAPC4 acts as a versatile bridge, interacting with Pol II and Pol III machinery differently, thanks to its dual-sided interaction motifs. Finally, using cross-linking mass spectrometry combined with AlphaFold structural simulations, the researchers obtained detailed maps of the SNAPc subunits SNAPC2 and SNAPC5, showing their roles in stabilising the complex and regulating DNA binding.
These findings clarify the mechanics of snRNA production at specialised snRNA genes and lay the foundation for future research into transcription regulation and its implications in health and disease. Furthermore, these results expand our understanding of how cells precisely manage complex molecular tasks, offering insights into fundamental biology and potential medical advances.
Shah, S.Z., Perry, T.N., Graziadei, A. et al. Structural insights into distinct mechanisms of RNA polymerase II and III recruitment to snRNA promoters. Nat Commun 16, 141 (2025). https://doi.org/10.1038/s41467-024-55553-8



