
Erdmann Group
Liquid Phase Separated Compartments and the Need for Subcellular Targeting
Compartmentalization is a characteristic feature of cellular organization. It serves to separate incompatible chemical reactions, enables the creation of specialized micro-environments, and can make chemical reactions more efficient by limiting the molecular actors involved to partial volumes. Traditionally, it refers to membrane-bound compartments such as the endoplasmic reticulum (ER), the Golgi apparatus or the mitochondria. Membranes in these classical organelles not only represent separating interfaces, but can also serve as two-dimensional platforms for the organization of reaction pathways.
In recent years, a new concept of cytoplasmic organization has emerged: membrane-less compartments resulting from liquid-liquid phase separation (LLPS). Examples for such compartments in eukaryotes include Cajal bodies and the nucleolus. There is growing evidence that LLPS is of great importance for understanding the pathology of several communicable diseases, neurodegenerative disorders and aging.
Consequently, there is a need for methods that allow studying LLPS within intact cells (‘in situ’).
To address this need, we use a combination of cryo-fluorescence microscopy (FLM), cryo-lift out (LO), cryo-focused ion beam (FIB) milling, and cryo-electron tomography (ET) to study liquid-liquid phase separated compartments inside vitrified cells and tissue. We constantly strife to improve the accuracy of this pipeline by developing both new soft- and hardware.
Recent and Ongoing Studies in our Group:
Using the 3D correlative FIB approach (Fig. 1), we are investigating the ultrastructure of both naturally occurring, as well liquid-liquid phase separated compartments formed during disease processes. Recently, we have provided insights into the organization of the nucleolus, an example of constitutive LLPS in all eukaryotic cells and place of ribosome biogenesis. By working in algae, yeast and mammalian cells, we can follow the formation of the small as well as the large subunit over a broad range of the evolutionary tree, revealing new intermediates that may have been missed by single particle experiments.
In addition to the nucleolus, there are many more transient LLPS compartments inside of cells for which an ultrastructural description is still missing. In a collaboration with Florian Wilfling (MPI for Biophysics, Frankfurt), we recently discovered and described the ultrastructure of a new liquid-phase separated compartment involved in clearing of failed endocytic events in S. cerevisiae. This LLPS, which we named ‘END’ for endocytic protein deposit, helps to buffer the proteins of early endocytosis and surrenders them to the autophagic machinery if needed.
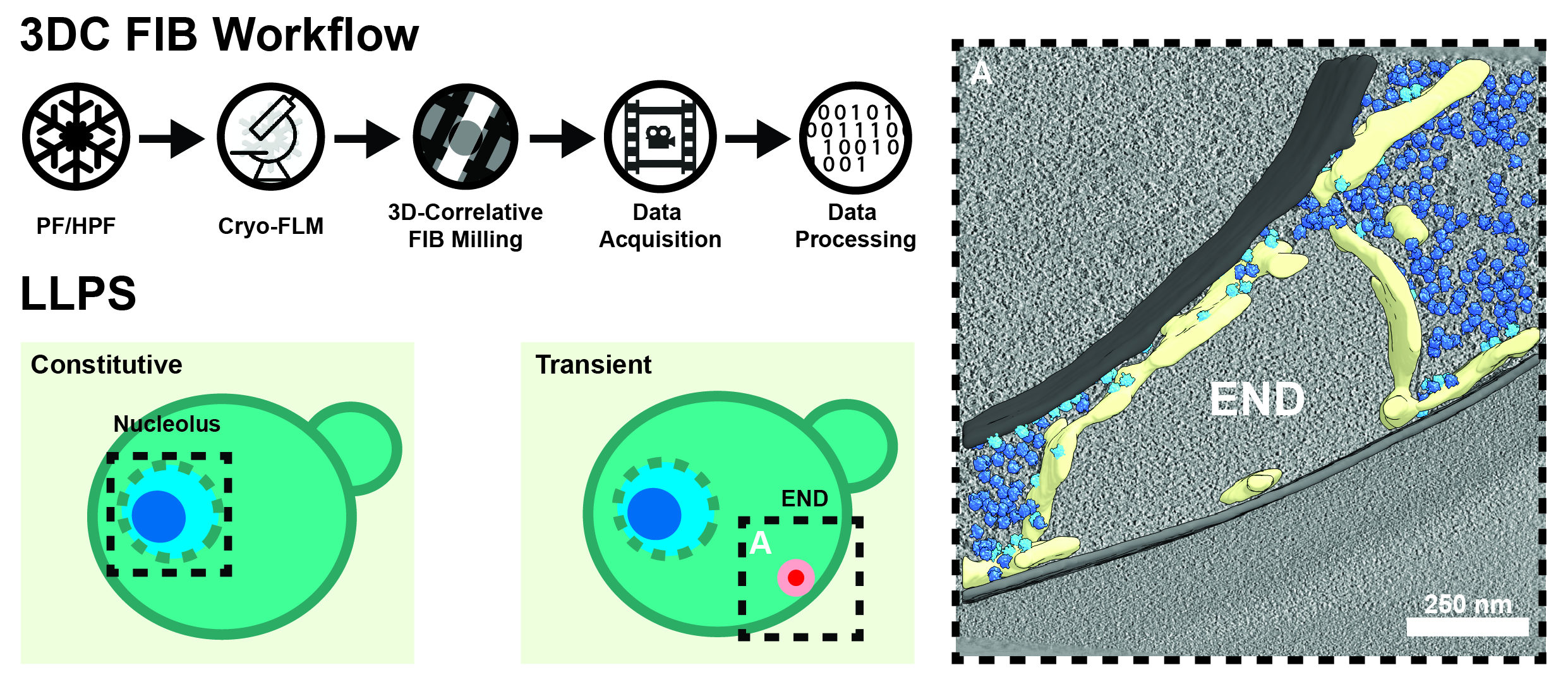 Figure 1. 3D-Correlative (3DC) Focused Ion Beam (FIB) Milling. Starting from either plunge-frozen (PF), or high-pressure frozen (HPF) samples, we perform cryogenic confocal fluorescence light microscopy (FLM) to localize features of interest. Using this information, we target our lamellas during FIB milling before they are subjected to automated cryo-tomogram acquisition and processing. We have used this technique on a variety of LLP compartments already, including the nucleolus of the green alga C. reinhardtii and the endocytic protein deposit (END), which plays a critical role in the homeostasis of early endocytic proteins in S. cerevisiae (right; membranes = gray, ER = yellow, free ribosomes = dark blue, ER-associated ribosomes = light blue).
Figure 1. 3D-Correlative (3DC) Focused Ion Beam (FIB) Milling. Starting from either plunge-frozen (PF), or high-pressure frozen (HPF) samples, we perform cryogenic confocal fluorescence light microscopy (FLM) to localize features of interest. Using this information, we target our lamellas during FIB milling before they are subjected to automated cryo-tomogram acquisition and processing. We have used this technique on a variety of LLP compartments already, including the nucleolus of the green alga C. reinhardtii and the endocytic protein deposit (END), which plays a critical role in the homeostasis of early endocytic proteins in S. cerevisiae (right; membranes = gray, ER = yellow, free ribosomes = dark blue, ER-associated ribosomes = light blue).
Localizing and targeting subcellular structures such as liquid-liquid phase separated compartments however, is only the beginning of the story. Understanding their inner structure could potentially help elucidate their role in normal cell function and disease. When working in complex cellular environments such as tissue samples, however, establishing and confirming the identity of the cells being observed by TEM can be challenging – despite having cryo-FLM at our disposal. For example, how to differentiate between gut, nerve and reproductive cells in high pressure frozen C. elegans samples at the TEM level (Fig. 2, left)?
There is clearly a need for both cryo-markers and tags that can be used to establish a) cellular identity and b) localize proteins, which are too small or too disordered to be recognized directly by cryo-TEM. In order to address cellular identity, we use naturally occurring (bacterial encapsulins, EncA) and de-novo designed proteins complexes, which can be expressed under tissue-specific promotors. These mega-Dalton complexes can easily be detected even in highly crowded environments such as mouse hippocampal synapses (Fig. 2, middle). However, EncAs cannot be used as labels for individual proteins due to their size and multiplicity of binding. Therefore, we are developing tags based on small artificial proteins, which can be directly linked to targets of interest and thus can relay their position via a characteristic shape. Unlike the EncA markers, much smaller, but at the same time still detectable and uniquely shaped protein designs are needed for this. A challenging task for our team and collaborators!
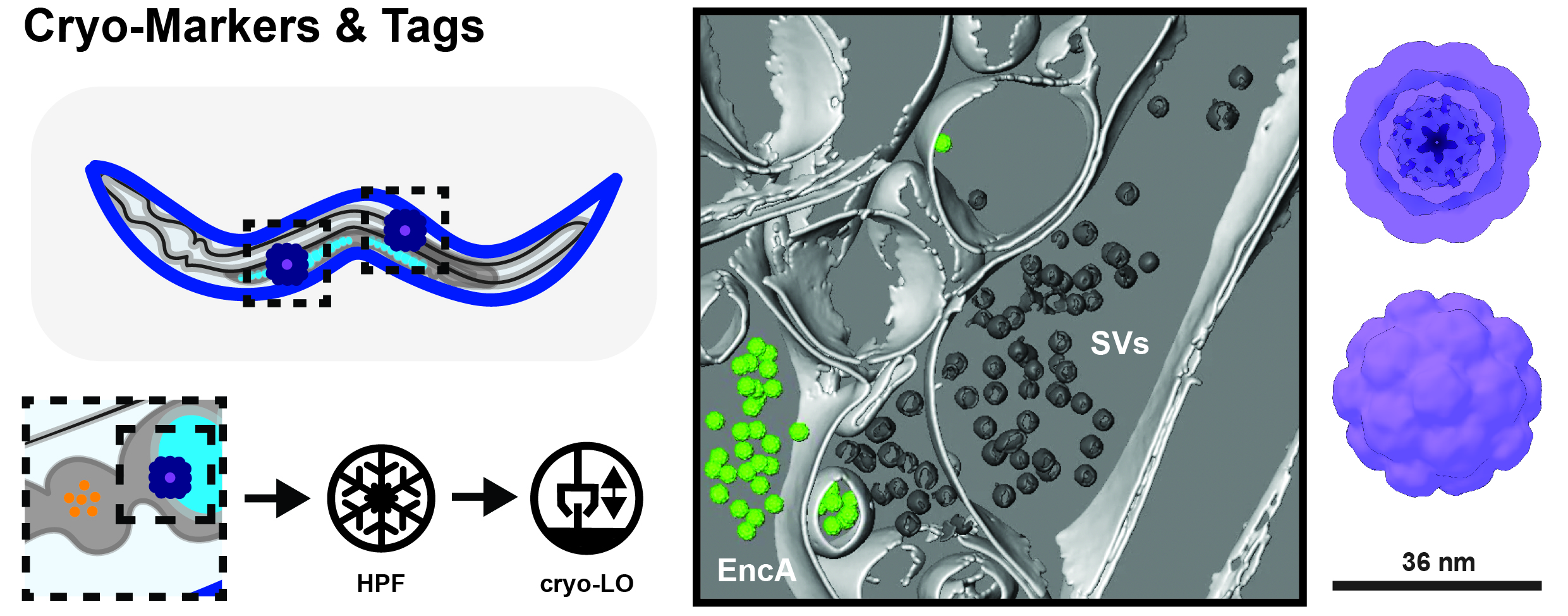
Figure 2. Cryo Markers and Tags for in-situ Work. Establishing cellular identity in cryo-lift out (LO) samples can be challenging since there are not many endogenous features to go by (left). Hence, we are using non-reactive protein complexes such as encapsulins (EncAs), which can be expressed under tissue-specific promoters. These can easily be detected e.g. at synapses of dissociated mouse hippocampal neurons (middle; membranes = light gray; synaptic vesicles = dark gray; EncA = green).
Group members
-
 Laura Bonanomi
Laura Bonanomi
PhD Student -
 Claire Overly Cottom
Claire Overly Cottom
Postdoc -
 Philipp Sebastian Erdmann
Philipp Sebastian Erdmann
Research Group Leader -
 Andrea Ghisleni
Andrea Ghisleni
Senior Technician -
 Jasmine Nguyen
Jasmine Nguyen
PhD Student -
 Alessandra Palumbo
Alessandra Palumbo
Undergraduate Intern -
 Gaia Perone
Gaia Perone
PhD Student -
 Uriel Petralia
Uriel Petralia
PhD Student -
 Janek Uhlich
Janek Uhlich
Undergraduate Intern -
 Jessica Winter
Jessica Winter
Scientific Visitor
