A new approach to investigate the impact of retrotransposon recombination on human genome
Recombination of retrotransposons is known to affect genome stability and evolution, yet the significance of this process and its impact on the genome of human somatic cells are poorly understood. Giovanni Pascarella and Piero Carninci set up a new framework enabling comprehensive studies of retrotransposon recombination-driven genomic diversity in healthy and diseased tissues.
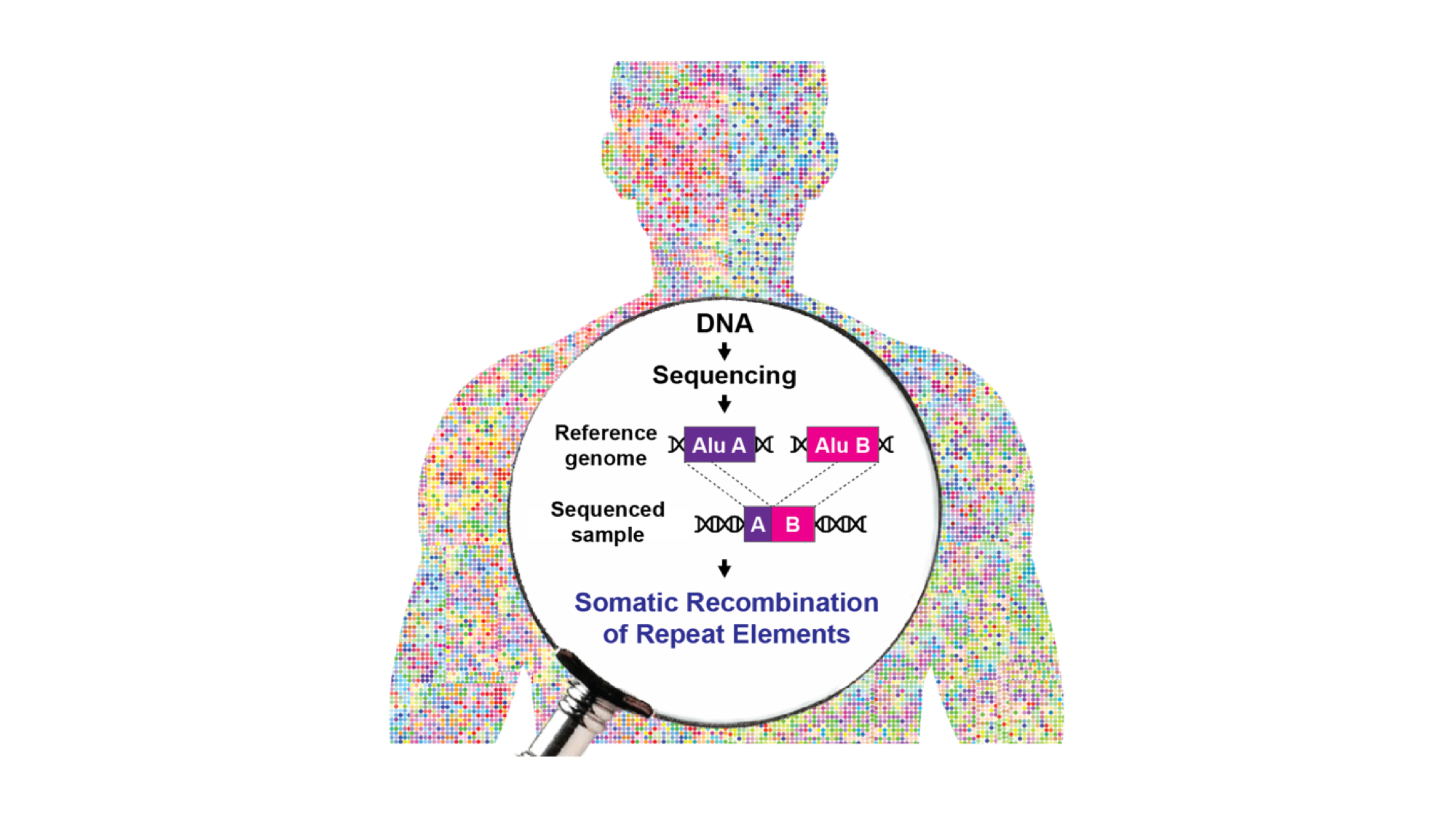
The DNA confined within the nucleus in each cell of our body is constantly subjected to endogenous and exogenous insults that can result in double strand breaks (DSBs). These DNA lesions endanger the fitness of the genome, and therefore they need to be repaired promptly to avoid the triggering of apoptotic pathways. A major pathway of DNA DSBs repair is homologous recombination, which can happen i) between two identical loci on sister chromatids or homologous chromosomes, leading to neutral resolution of the break, or ii) between non-allelic loci (non-allelic homologous recombination, NAHR). This latter process can create genetic diversity and contribute to genome evolution, but it can also be detrimental for the organism. The human genome hosts millions of copies of retrotransposons or transposable elements, LINE-1 (L1) and Alu being the most abundant and accounting for approximately one-third of the genomic sequence. Due to the high number of copies and high identity, these transposable elements have been often found involved in NAHR events leading to different types of DNA rearrangements in cancer and genetic disorders. Yet, the impact of NAHR on somatic genomic variation and the contribution of different retrotransposons to NAHR-dependent genome rearrangement remain ill defined.
To address these issues, Piero Carninci – Head of the Genomics Research Centre at Human Technopole, Team Leader for Laboratory for Transcriptome Technology, and Deputy Centre Director at the RIKEN Centre for Integrative Medical Sciences (IMS) in Yokohama (Japan) – and Giovanni Pascarella – research scientist at IMS – combined an optimised protocol for the enrichment of retrotransposons sequences (“capture-seq”) with a novel bioinformatic analytical workflow (TE-reX) to comprehensively investigate somatic NAHR events in Alu and L1 elements in post-mortem tissues of different embryonic derivation, including liver, kidney, and cortical brain regions from ten neurotypical donors. The results of the study are now published in Cell.
The researchers found that the number of NAHR events associated with Alu and/or L1 recombination was higher in the liver and kidney than in brain tissue. Of note, the brain showed high rates of intra-chromosomal NAHR events and recombination among repeat elements in close proximity, preferentially occurring in the shorter human chromosomes. In addition, intra-chromosomal recombination of close inverted repeats represented a source of genomic instability in a tissue-dependent manner. Furthermore, L1 and Alu were enriched at human centrosomes, the latter also found as hotspots in oncogenes and tumour suppressor genes in the COSMIC database. But what are the origin and physiological role of tissue-specific recombination of Alu and L1? Pascarella et al. addressed this question analysing somatic NAHR events in vitro, inducing pluripotent stem cells (iPSCs) to differentiate into neurons (iNEUs). In agreement with the results obtained in brain tissue, iNEUs displayed higher recombination rates of proximal Alu and L1 pairs and a marked inverse correlation between chromosome length and intra-chromosomal recombination rates compared to iPSCs. This suggests that recombination of Alu and L1 sequences may be linked to chromatin rearrangements during early neuronal differentiation and brain development.
“Our analyses of recombination in iPSC and differentiated neurons suggest that the specific recombination profiles observed in post-mortem samples may be generated in progenitor cells during early developmental stages. It is nevertheless an open question whether tissue- specific recombination profiles are a cause or consequence of cell-fate determination” says Giovanni Pascarella, first author of the study.
Next, the team investigated tissue-specific NAHR of Alu and L1 in two pathological conditions, sporadic Parkinson’s (PD) and Alzheimer’s (AD) disease, by profiling somatic NAHR of Alu and L1 in post-mortem tissue samples from PD and AD patients. These analyses revealed that NAHR events are enriched in PD- and AD-associated genes in a disease- and tissue-specific fashion.
“The breakpoints of somatic NAHR causing intra-chromosomal rearrangements are enriched in a tissue- and disease-specific fashion in PD- and AD-associated genes and this may be consequential to complex alterations of both DNA damage and DNA repair in neurodegeneration. More work will be required to understand the significance and impact of neurodegeneration-specific alterations of NAHR profiles” explains Piero Carninci, Principal Investigator and co-corresponding author of the study.
In summary, Pascarella and Carninci show for the first time that global somatic NAHR of Alu and L1 occurs in physiological conditions in a tissue-dependent manner and that NAHR hotspots are enriched at centrosomes and in both oncogenes and tumor suppressor genes. Moreover, their results suggest that somatic NAHR events may trigger early developmental differentiation, as well as the transition from healthy to pathological states in human cells and tissues. While the reason why the human genome carries such a high number of transposable elements is still under debate, these findings deepen our understanding on the complex tissue-dependent dynamics of somatic NAHR and their impact on the structure and function of human genome.