Cryo-EM sheds light on the specificity of retrotransposon integration
Retrotransposons or transposable elements (TEs) are parts of eukaryotic DNA that can replicate and paste themselves into different locations in the genome. The process involves reverse transcription converting RNA into DNA, which is then inserted into the host genome. This “copy-and-paste” mechanism is shared with a class of viruses called retroviruses but does not lead to the formation of infectious particles that may infect other cells. Yet, retrotransposon replication and insertion are potentially harmful as they could interrupt the coding sequence of a gene or create sites where DNA can recombine, thereby inducing new mutations in the genome.
Long terminal repeat (LTR) retrotransposons are the most abundant class of TEs in the eukaryotic genome. Ty3/Gypsy is a well-characterized LTR in budding yeast Saccharomyces cerevisiae, which is known to insert upstream of transfer RNA (tRNA) genes that are essential for protein synthesis. General transcription factors TFIIIB and TFIIIC mediate Ty3 insertion at tRNA genes, conferring targeting specificity and directionality of integration, respectively. However, little is known about how such a specificity is achieved.
Alessandro Vannini, Head of the Structural Biology Research Centre at Human Technopole, and his team used cryo-electron microscopy (cryo-EM), an advanced imaging technique, to characterize the interaction of Ty3, TFIIIB, and TFIIIC with the target tRNA genes and reveal the molecular mechanisms underlying the site-specificity of Ty3 integration. The results of this research have been published in Nature Communications.
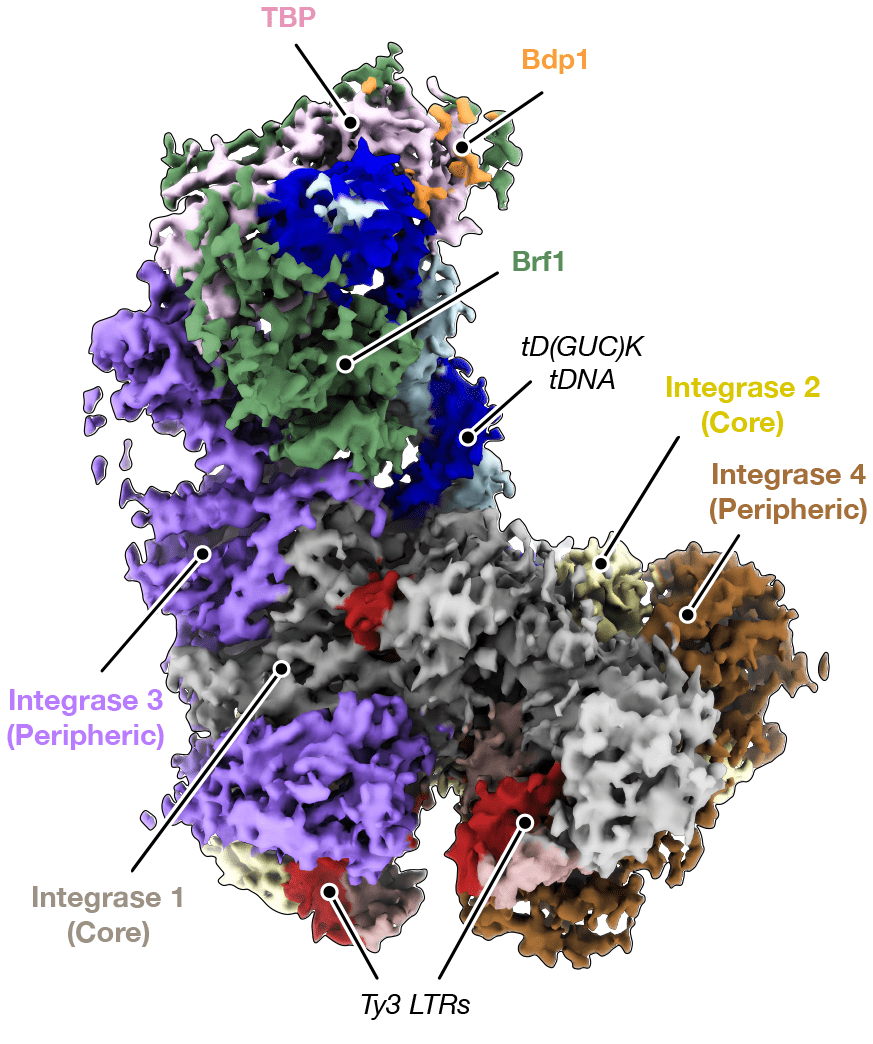
The researchers purified the Ty3 retrotransposon machinery – consisting of a reverse transcriptase and an integrase, hereinafter referred to as intasome – and reconstituted the Ty3 intasome engaged with TFIIIB alone or in complex with TFIIIB and target DNA using cryo-EM.
They found that the C-terminal domain (CTD) and a previously uncharacterised chromodomain (CHD) of Ty3 integrase, specified the position of Ty3 gene insertion upstream of the target tRNA gene promoter. Furthermore, a long linker region between the CTD and the catalytic core domain (CCD) of Ty3 integrase turned out to be required for the recruitment of Ty3 intasome by TFIIIB. Notably, the binding mode of Ty3 intasome to TFIIIB resembled that of the human immunodeficiency virus (HIV) intasome to the lens epithelium-derived growth factor (LEDGF/p75) gene. This parallel suggests that, despite being evolutionarily distant, HIV and Ty3 intasomes use conserved regions at their periphery to target host DNA.
To investigate how Ty3 retrotransposon integration takes place, the researchers compared the cryo-EM structures of RNA Polymerase III (Pol III) – responsible for tRNA gene transcription – and Ty3 intasome recruited by TFIIIB to a Pol III-transcribed gene and showed that their binding to the target DNA requires equivalent regions, which prevents co-occurrence of tRNA transcription and Ty3 integration.
In summary, Vannini and his group revealed the structural basis underlying the specificity of Ty3 integration at Pol III-transcribed genes and showed how these features are conserved in evolutionarily distant TEs.



