Clearer microscopy images in spatial biology
An international team of scientists led by HT researchers Magda Bienko and Nicola Crosetto developed an open-source software for deconvolution of widefield fluorescence microscopy image stacks and large tissue scans. This new tool increases the information obtained with fluorescence microscopy-based spatial omic methods.
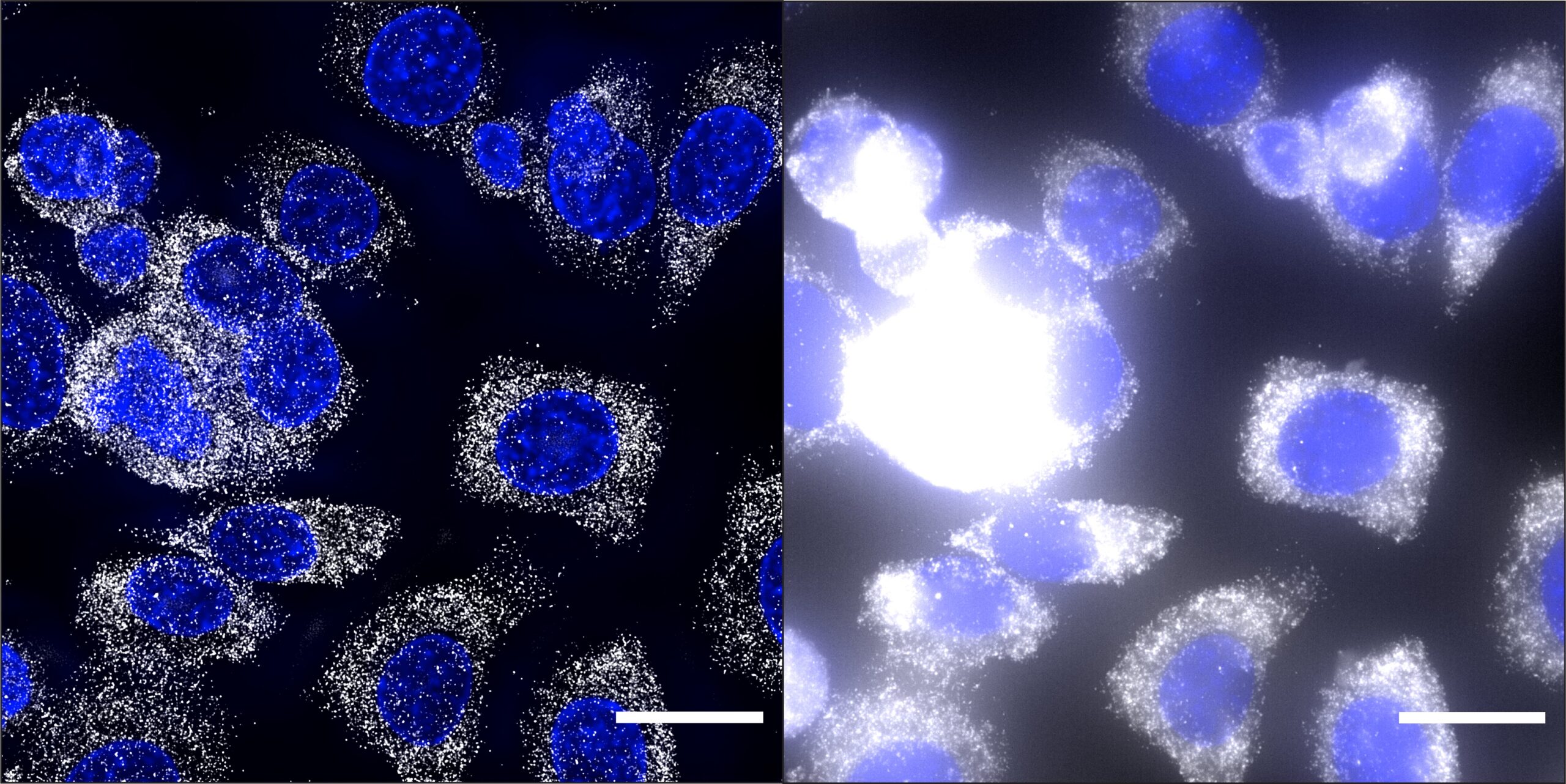
“Resolution” is a term that microscopists use to indicate the ability to distinguish details in an imaged sample. The higher the resolution of a micrograph, the clearer and sharper its fine elements. Image resolution strictly depends on the characteristics of a microscope, such as the type of lens, the wavelength of the light used for imaging and the removal of out-of-focus signal. Confocal and super-resolution microscopes ensure high resolution but are expensive and require highly trained operators.
Deconvolution is an image-processing technique that increases the contrast and resolution of images, such as DNA loci, RNA molecules, or proteins in single cells within tissues and organs, using conventional optical microscopes and fluorescence microscopy-based spatial omic methods. However, commercially available deconvolution software have several limitations, ranging from high costs, the inability to process large image datasets and crowded images, to the need for extensive hands-on training and powerful computers.
To overcome these drawbacks, Magda Bienko and Nicola Crosetto, researchers at the Genomics Research Centre at Human Technopole, together with an international team of researchers from Sweden, Spain, and the USA, developed the open assay and easy-to-operate deconvolution software Deconwolf (DW).
The results of the research are published in Nature Methods.
The team tested Deconwolf on fluorescence microscopy images displaying (near-)diffraction limited fluorescence dots (DNA and RNA FISH images) or crowded spatial omics-derived images such as in situ spatial transcriptomics. They showed that Deconwolf improved the resolution and amount of information in images obtained with these high-throughput methods. In addition, it outperformed two among the most used commercially available deconvolution software.
Deconwolf can be freely downloaded from https://deconwolf.fht.org/ (web development by Riccardo De Lucia, Human Technopole Computational Biology) and runs on standard laptops.
“DW represents a potentially transformative tool that can be adopted across many fields of life sciences and biomedicine to increase the amount of biological information retrieved from imaging data generated by techniques that rely on fluorescence detection using widefield microscopy and imaging-based spatially resolved omic methods that generate (near-)diffraction limited signals. We therefore anticipate that DW will democratise the use of deconvolution in bioimaging and find numerous applications in research and diagnostics”, the authors say.
Wernersson, E., Gelali, E., Girelli, G. et al. Deconwolf enables high-performance deconvolution of widefield fluorescence microscopy images. Nat Methods (2024). https://doi.org/10.1038/s41592-024-02294-7