Microcephalin 1 regulates chromosomal DNA condensation
The nucleus of eukaryotic cells contains the genome of an organism, which consists of chromosomal DNA arranged in well-organised protein-DNA structures. To proliferate, cells first duplicate their chromosomal DNA and then distribute it evenly between to daughter cells by means of a multistep process referred to as cell cycle. During cell division or mitosis, the last step of this cycle, the chromosomal DNA becomes tightly condensed and acquires a X-shaped structure with two arms or sister chromatids, which are then pulled in opposite direction and segregated between daughter cells. Mitotic condensation of chromosomal DNA is necessary for the correct segregation of sister chromatids.
Cohesin and condensin are protein complexes that promote changes in chromosomal DNA topology during interphase – the preparation phase for cell division – and mitosis, respectively, by extruding DNA loops. Cohesin release factor WAPL is known to block cohesin association with chromosomal DNA thereby modulating cohesin’s processivity. However, little is known about how condensin’s activity and function are regulated in mammalian cells. Condensin II, one of the two mammalian condensin complexes, has been previously shown to be activated during mitosis. However, the characterization of cells from patients affected by congenital primary microcephaly suggests that condensin II may be active in interphase and induce premature chromosomal DNA condensation.
Alessandro Vannini, Head of the Structural Biology Research Centre at Human Technopole, and colleagues have now shown that Microcephalin 1 (MCPH1), a gene expressed during foetal brain development, the mutation of which leads to primary microcephaly, prevents stable association of condensin with chromosomal DNA. In turn, this reduces processive loop extrusion by condensin processivity and affects the organisation of chromosomal DNA. These results have been published in eLife.
By deleting Mcph1 gene in mouse embryonic cells, Vannini et al. found that the loss of Mcph1 led to chromosome condensation during interphase and condensin II’s stable association with interphase chromosomal DNA. A combination of imaging and biochemical approaches revealed that MCPH1 directly binds a short Linear Motif (SLiM) located within the central domain of condensin II and that the SLiM was essential for condensin II to inhibit premature condensation of chromosomal DNA during interphase.
Next, the authors increased MCPH1 levels in mouse oocytes, specialized germ cells involved in reproduction that divide via a special type of cell division called meiosis. In this way, they could inhibit the association of condensin II with chromosomes during interphase but not meiosis, suggesting a cell cycle-dependent role for MCPH1 in regulating chromosomal DNA condensation.
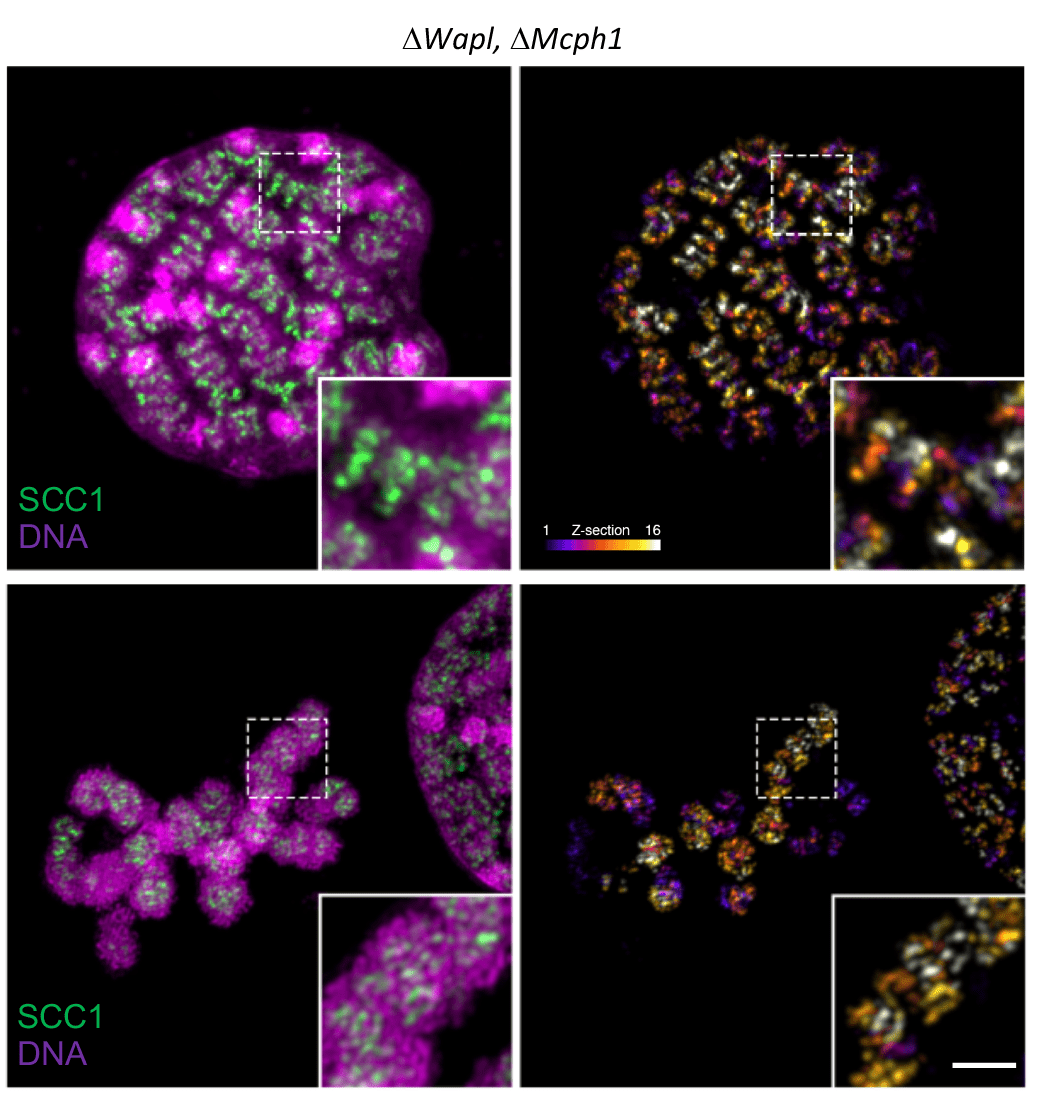
Finally, the researchers investigated the architecture of interphase chromosomal DNA in mouse cells lacking Wapl or Mcph1, or both. They observed that the loss of WAPL and that of MCPH1 induced the formation of thread-like structures (vermicelli) and soft spherical or “gumball” chromosomes, respectively. The simultaneous deletion of Wapl and Mcph1 led to the formation of chromosomes with a solenoid morphology, suggesting that the activities of cohesin and condensin complexes must be tightly regulated to ensure correct chromatid formation and segregation.
In summary, Vannini and colleagues suggest that MCPH1 regulates the association of condesin II with chromosomal DNA during interphase via a mechanism resembling cohesin regulation by WAPL. Interestingly, these findings also show that condesin II has cell cycle phase-specific regulatory mechanisms. A better understanding how MCPH1/condesin II and WAPL/cohesin jointly regulate chromatid formation and segregation holds promise to shed new light on how loop extrusion creates chromosomes.