Researchers map a fundamental enzyme
For the first time, scientists have “photographed” and mapped human RNA polymerase III, a fundamental enzyme for the life of all cells whose mutations can cause hypersensitivity to viruses with severe consequences to our health. This result was obtained by researchers from the Institute of Cancer Research in London and the University of Regensburg (Germany), led by Alessandro Vannini, Head of Structural Biology Research Centre at Human Technopole.
This discovery could have important and practical implications: understanding the mutations of this molecule could allow to develop tools and technologies to contrast its negative effects, such as hypersensitivity to viruses as well we neurological development disorders and types of tumours where this molecule is “excessively” active.
The results were published today by the prestigious scientific journal Nature Communications.
The researchers’ study is the first complete and high-resolution representation of human RNA polymerase III. This macromolecule allows to transfer genetic information from DNA to RNA in most living beings, from plants to animals and humans. Until today the detailed structure of RNA polymerase III was available only for yeast (Saccharomyces cerevisiae) while its human counterpart remained unexplored.
Researchers were finally able to reach this result thanks to cryo-electron microscopy, a technology allowing scientists to see molecules at atomic scale and which was considered so advanced and revolutionary to deserve a Nobel Prize for Chemistry in 2017.
Alessandro Vannini, Head of the Structural Biology Research Centre at Human Technopole commented: “Today’s study allows us to identify which are the mutations connected to numerous diseases and where they are positioned. Over the last few years, a large number of genetic mutations in RNA Polymerase III have been identified as responsible for hypersensitivity to viral infections and alterations to a proper development of the central nervous system as well as severe neurodegenerative diseases. In addition, this enzyme is “hyperactive” in cancer: understanding its detailed structure can be of great help in developing targeted therapies”.
The study
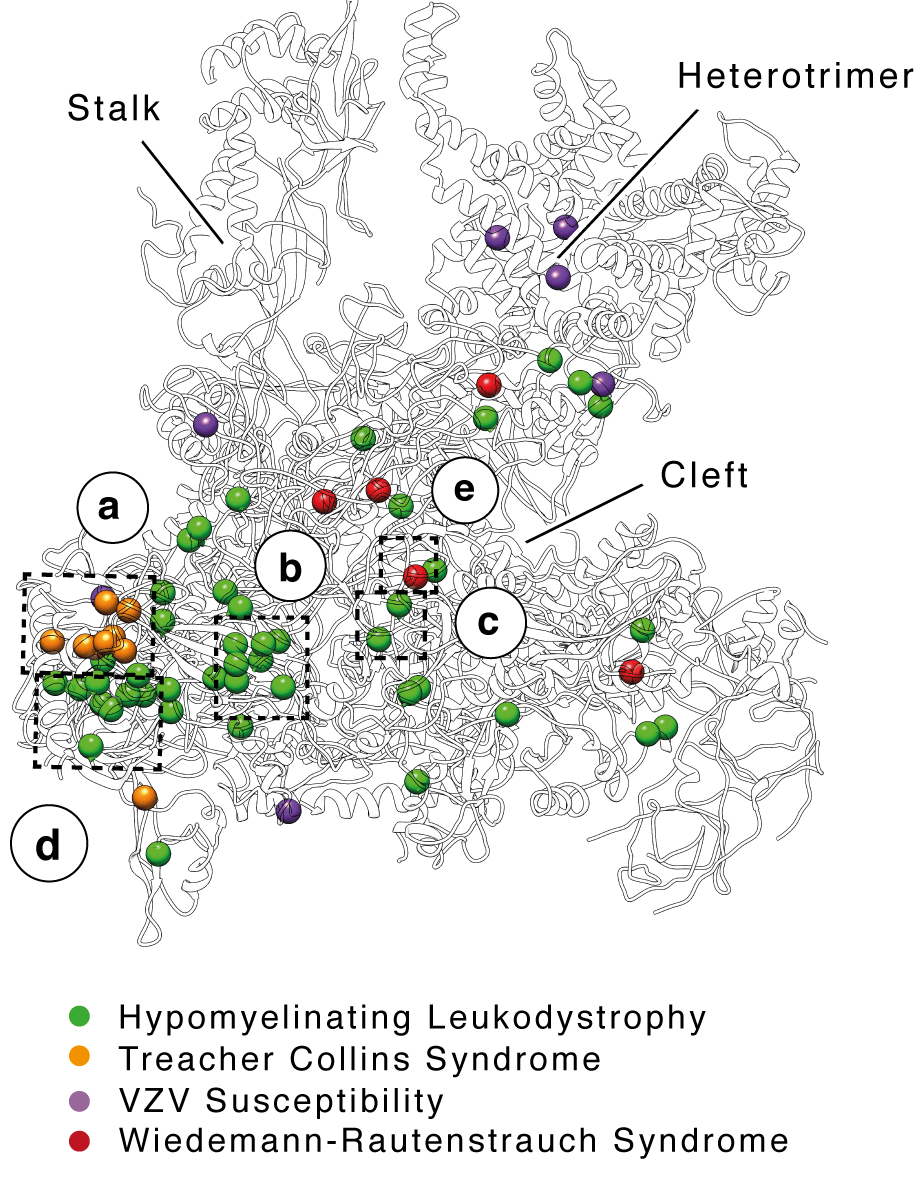
Researches were able to extract RNA polymerase III from human cells thanks to advanced genetic engineering techniques, including CRISPR/Cas9, the technology awarded this year with the Nobel Prize for Chemistry. Once isolated, the macromolecule was “photographed” thanks to the use of cryoelectron microscopy. Researchers where then able to obtain the first high resolution image of its structure. This allowed them to map over 85% of its genetic mutations with a high level of precision as well as to study how these interact with other structural elements at molecular level.
Researchers observed that these mutations cause neurodevelopmental defects, such as hypomyelinating leukodystrophy, Treacher-Collins syndrome or Wiedemann-Rautenstrauch syndrome (WRS), cluster together around central points in the enzyme, influencing its stability and biogenesis. On the other hand, the alterations which influence the hypersensitivity to viruses are positioned on the outside, closer to the regions where the molecule recognises DNA of viral origin.
RNA polymerase
A specific group made up of three enzymes known as RNA polymerases is necessary to allow genetic information within a DNA molecule to be copied faithfully. These enzymes allow the transcription of genetic information from DNA to RNA. The latter is then transported in other parts of the cell to absolve important functions such as the production of proteins, which is essential to life.
RNA polymerase III is fundamental to determine cellular growth and lifespan among eukaryotes, meaning all those living beings whose cells have a nucleus, from plants to animals and humans. Within eukaryotes, this macromolecule shows a high level of conservation both in terms of composition of its parts, as well as of homology in the sequence of its individual components. A notable exception is sub-unit RPC5, which in animals is different and whose function is currently unknown. The current study reveals that this sub-unit is important for the correct assembly of the enzyme, suggesting a finer level of regulation in animals.
Human Technopole Structural Biology Research Centre
The Centre represents one of the five highly relevant and complimentary research areas of Human Technopole’s initial activity. It is led by Prof. Alessandro Vannini. The Centre aims to gain a precise knowledge of the structure of macromolecules, which is essential to understand how they function. Human Technopole’s experimental genomics and neurogenomics activities will generate information on many novel genes and proteins, including targets for the treatment of diseases. In this context, Human Technopole’s structural biologists will seek to characterise the 3D structures of these targets to understand their functions and modes of action, as well as to develop new drugs. Experimental research activity will be supported by state-of-the-art cryo-electron microscopy technology to support high resolution molecular structure determination. Over the past ten years cryo-electron microscopy (CryoEM) has emerged as a powerful tool to study structure and function of macromolecules in great detail. Today Human Technopole is setting up a facility with five microscopes, making it one of the most advanced facilities in Europe.
*************************
Nature Communications, 17 December 2020
Structure of human RNA Polymerase III
Ewan Phillip Ramsay (1,†), Guillermo Abascal-Palacios (1,†), Julia L. Daiß (2,†), Helen King (1), Jerome Gouge (1), Michael Pilsl (2), Fabienne Beuron (1), Edward Morris (1), Philip Gunkel (3), Christoph Engel (2,*) e Alessandro Vannini (1,4,*)
- Division of Structural Biology, The Institute of Cancer Research, London SW7 3RP, United Kingdom.
- Regensburg Center for Biochemistry, University of Regensburg, 93053 Regensburg, Germany
- Max Planck Institute for Biophysical Chemistry, Research Group Nuclear Architecture, 37077 Göttingen, Germany
- Fondazione Human Technopole, Structural Biology Research Centre, 20157 Milan, Italy
†All authors contributed in equal measure to the study