Ultrastructure Expansion Microscopy details Chlamy native organisation
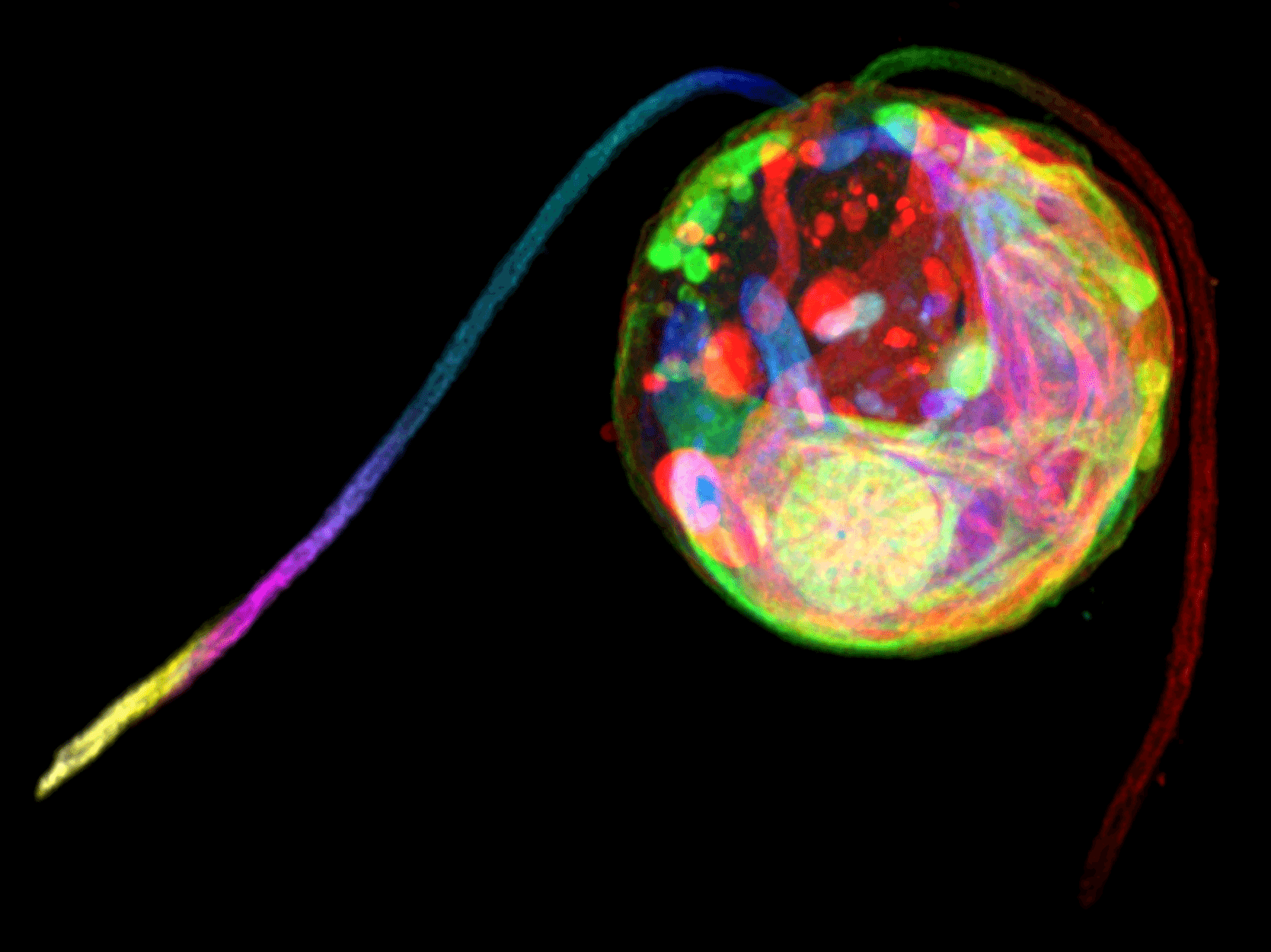
The Pigino Group developed a detailed protocol to obtain super-resolution images of the green alga Chlamydomonas reinhardtii by using conventional optical light microscopes.
The advent of super-resolution imaging techniques coupled with fluorescence microscopy allowed the visualisation of a vast array of biological samples with a resolution higher than that imposed by the light diffraction limit. However, super-resolution microscopes are usually expensive instruments with complicated setups and require experienced personnel for sample preparation and image acquisition. Expansion Microscopy (ExM) has recently been developed to overcome these limitations and uses swellable hydrogels to obtain up to fourfold isotropic expansion of the sample before imaging with a confocal microscope.
Gaia Pigino, Associate Head of the Structural Biology Research Centre at Human Technopole, Nikolai Klena and Giovanni Maltinti, members of the Pigino Group, in collaboration with Paul Guichard and Virginie Hamel at the University of Geneve, developed an ExM-based protocol to visualise the three-dimensional (3D) ultrastructural organisation of Chlamydomonas reinhardtii (Ultrastructure Expansion Microscopy, U-ExM). The protocol is now published in the open-access journal Bio-protocol and made available to the research community.
Chlamydomonas is a single-cell model organism widely used to investigate the molecular mechanisms of cilia and flagella motility. In this protocol, the researchers compare different fixation and staining procedures and provide instructions on how to embed the sample in a hydrogel and acquire images with a confocal microscope as well as how to analyse them.
The Pigino Group has already used this protocol to study the assembly of intraflagellar transport trains (IFT) at the Chlamydomonas ciliary base1, thus showing that this new procedure will be instrumental for researchers to perform super-resolution analysis of target proteins in a native 3D environment without the need for expensive super-resolution microscopes and specific training.
1 van den Hoek et al. (2022) In situ architecture of the ciliary base reveals the stepwise assembly of intraflagellar transport trains. Science