Vannini Group
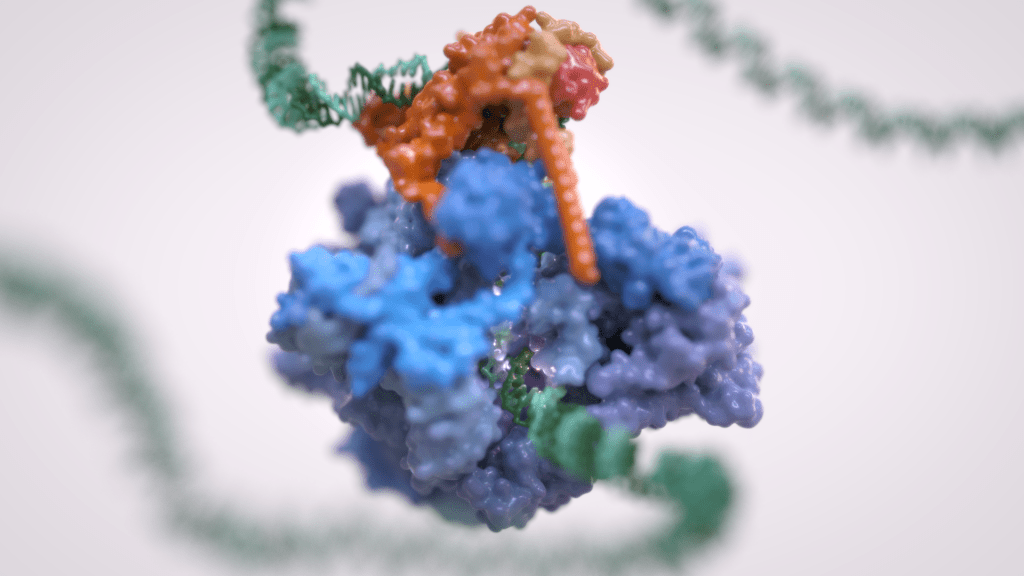
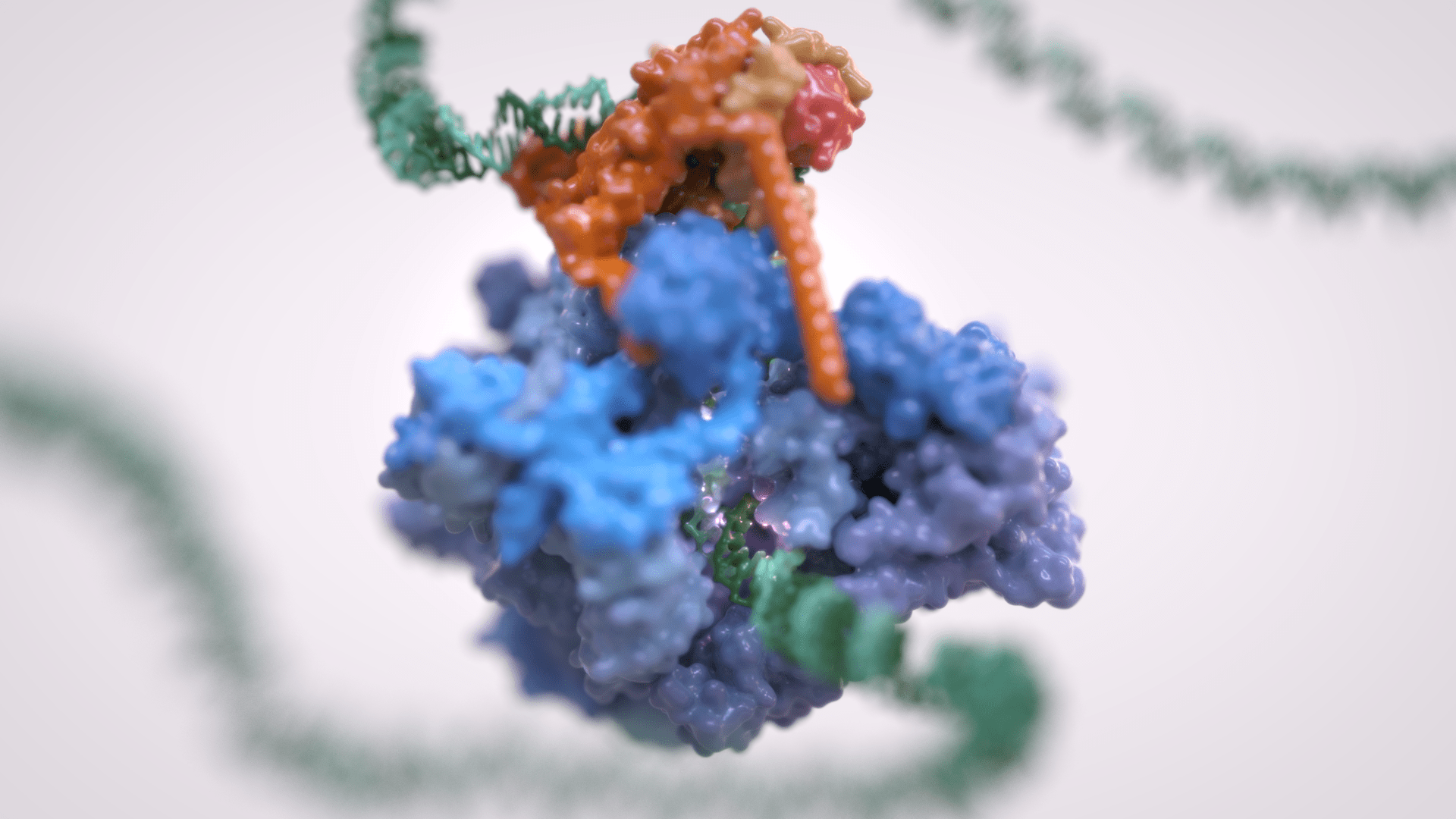
Gene transcription is the first step that controls the expression of the genetic information encoded in a genome and ultimately underlies cell differentiation and organism development. Eukaryotic gene transcription occurs in the context of highly structured and organised genomes and acts as a coordinator of numerous events co-occurring in the nucleus. Eukaryotic transcription relies on three different RNA polymerases: RNA polymerase I (Pol I) transcribes ribosomal RNA, RNA polymerase II (Pol II) synthesizes messenger RNAs and RNA polymerase III (Pol III) produces short and non-translated RNAs, including the entire pool of tRNAs, which are essential for cell growth.
For a long time, it was assumed that only Pol II was regulated whereas Pol I and Pol III did not require such control. However, it is now clear that RNA polymerase III transcription is tightly regulated and a determinant of organismal growth. Pol III deregulation is observed in many forms of cancer and Pol III genetic mutations cause severe neurodegenerative diseases.
Furthermore, Pol III and its associated factors play a paramount role into genome structure and organisation. These “extra-transcriptional roles” are carried out throughout interactions with other cellular components such as retroelement transposition machineries, Structural Maintenance of Chromosome (SMC) complexes and specific chromatin remodellers.
The Vannini Group employs an Integrative Structural Biology approach, combining cutting-edge cryo-EM analysis, x-ray diffraction data, cross-linking and native mass-spectrometry. We integrate the structural data with molecular and cellular biology techniques in order to obtain a comprehensive view of these fundamental processes and how their mis-regulation can lead to cancer and neurodegenerative diseases.
Group members
-
 Alessandro Vannini
Alessandro Vannini
Head of Structural Biology Research Centre -
 Alessandro Borsellini
Alessandro Borsellini
Postdoc -
 Giacomo Ettore Casale
Giacomo Ettore Casale
Postdoc -
 Valentina Cecatiello
Valentina Cecatiello
Senior Technician -
 Sebastian Chamera
Sebastian Chamera
Postdoc -
 Fabiola Iommazzo
Fabiola Iommazzo
PhD Student -
 Thomas Noé Perry
Thomas Noé Perry
Postdoc -
 Fabio Pessina
Fabio Pessina
Senior Technician -
 Mariavittoria Pizzinga
Mariavittoria Pizzinga
Postdoc -
 Ewan Ramsay
Ewan Ramsay
Senior Staff Scientist -
 Ankit Roy
Ankit Roy
PhD Student -
 Syed Zawar Shah
Syed Zawar Shah
PhD Student
Publications
-
11/2022 - Nature Structural & Molecular Biology
Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II
RNA polymerase II (Pol II) carries out transcription of both protein-coding and non-coding genes. Whereas Pol II initiation at protein-coding genes has been studied in detail, Pol II initiation at non-coding genes, such as small nuclear RNA (snRNA) genes, is less well understood at the structural level. Here, we study Pol II initiation at snRNA […]
-
09/2022 - Life Sci Alliance
The human RNA polymerase I structure reveals an HMG-like docking domain specific to metazoans
Transcription of the ribosomal RNA precursor by RNA polymerase (Pol) I is a major determinant of cellular growth, and dysregulation is observed in many cancer types. Here, we present the purification of human Pol I from cells carrying a genomic GFP fusion on the largest subunit allowing the structural and functional analysis of the enzyme […]
-
12/2021 - Elife
MCPH1 inhibits Condensin II during interphase by regulating its SMC2-Kleisin interface
Dramatic change in chromosomal DNA morphology between interphase and mitosis is a defining features of the eukaryotic cell cycle. Two types of enzymes, namely cohesin and condensin confer the topology of chromosomal DNA by extruding DNA loops. While condensin normally configures chromosomes exclusively during mitosis, cohesin does so during interphase. The processivity of cohesin’s loop […]
-
11/2021 - Nature Communications
Structural basis of Ty3 retrotransposon integration at RNA Polymerase III-transcribed genes
Retrotransposons are endogenous elements that have the ability to mobilise their DNA between different locations in the host genome. The Ty3 retrotransposon integrates with an exquisite specificity in a narrow window upstream of RNA Polymerase (Pol) III-transcribed genes, representing a paradigm for harmless targeted integration. Here we present the cryo-EM reconstruction at 4.0 Å of […]
-
09/2021 - Molecular Cell
A small nucleosome from a weird virus with a fat genome
Valencia-Sánchez et al. (2021) and Liu et al. (2021) provide structural and biological insights about the existence and importance of a nucleosome-like particle in a family of giant viruses.