Vannini Group
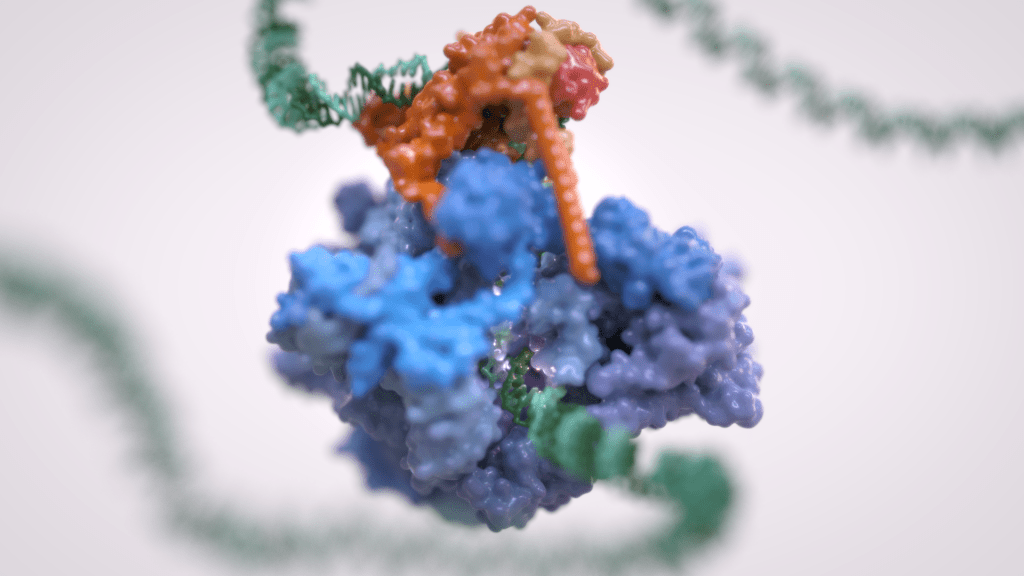
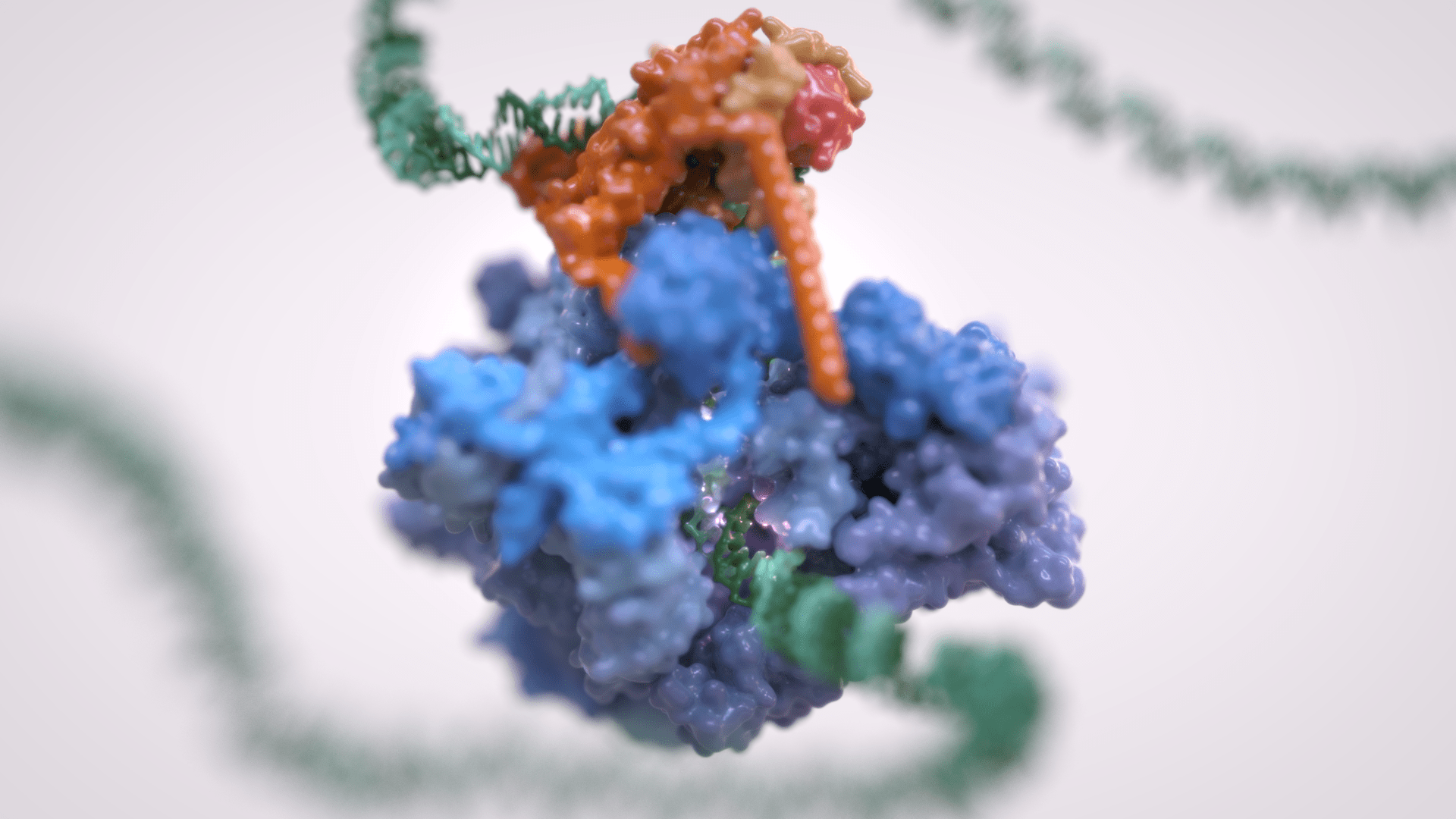
Gene transcription is the first step that controls the expression of the genetic information encoded in a genome and ultimately underlies cell differentiation and organism development. Eukaryotic gene transcription occurs in the context of highly structured and organised genomes and acts as a coordinator of numerous events co-occurring in the nucleus. Eukaryotic transcription relies on three different RNA polymerases: RNA polymerase I (Pol I) transcribes ribosomal RNA, RNA polymerase II (Pol II) synthesizes messenger RNAs and RNA polymerase III (Pol III) produces short and non-translated RNAs, including the entire pool of tRNAs, which are essential for cell growth.
For a long time, it was assumed that only Pol II was regulated whereas Pol I and Pol III did not require such control. However, it is now clear that RNA polymerase III transcription is tightly regulated and a determinant of organismal growth. Pol III deregulation is observed in many forms of cancer and Pol III genetic mutations cause severe neurodegenerative diseases.
Furthermore, Pol III and its associated factors play a paramount role into genome structure and organisation. These “extra-transcriptional roles” are carried out throughout interactions with other cellular components such as retroelement transposition machineries, Structural Maintenance of Chromosome (SMC) complexes and specific chromatin remodellers.
The Vannini Group employs an Integrative Structural Biology approach, combining cutting-edge cryo-EM analysis, x-ray diffraction data, cross-linking and native mass-spectrometry. We integrate the structural data with molecular and cellular biology techniques in order to obtain a comprehensive view of these fundamental processes and how their mis-regulation can lead to cancer and neurodegenerative diseases.
Group members
-
 Alessandro Vannini
Alessandro Vannini
Head of Structural Biology Research Centre -
 Alessandro Borsellini
Alessandro Borsellini
Postdoc -
 Giacomo Ettore Casale
Giacomo Ettore Casale
Postdoc -
 Valentina Cecatiello
Valentina Cecatiello
Senior Technician -
 Sebastian Chamera
Sebastian Chamera
Postdoc -
 Fabiola Iommazzo
Fabiola Iommazzo
PhD Student -
 Thomas Noé Perry
Thomas Noé Perry
Postdoc -
 Fabio Pessina
Fabio Pessina
Senior Technician -
 Mariavittoria Pizzinga
Mariavittoria Pizzinga
Postdoc -
 Ewan Ramsay
Ewan Ramsay
Senior Staff Scientist -
 Ankit Roy
Ankit Roy
PhD Student -
 Syed Zawar Shah
Syed Zawar Shah
PhD Student
Publications
-
06/2004 - J Biol Chem
Crystal structure of the quorum-sensing protein TraM and its interaction with the transcriptional regulator TraR
Transfer of the tumor-inducing plasmid in Agrobacterium tumefaciens is controlled by a quorum-sensing system whose main components are the transcriptional regulator TraR and its autoinducer. This system allows bacteria to synchronize infection of the host plant when a “quorum” of cells has been reached. TraM is an A. tumefaciens protein involved in the regulation of […]
-
02/2004 - Biochemistry
Mechanism of activation of human heparanase investigated by protein engineering
The aim of this study was to investigate the mechanism of activation of human heparanase, a key player in heparan sulfate degradation, thought to be involved in normal and pathologic cell migration processes. Active heparanase arises as a product of a series of proteolytic processing events. Upon removal of the signal peptide, the resulting, poorly […]
-
01/2004 - Acta Crystallogr D Biol Crystallogr
Crystallization and preliminary X-ray diffraction studies of the quorum-sensing regulator TraM from Agrobacterium tumefaciens
TraM is a 11.4 kDa protein involved in the control of the conjugal transfer of Agrobacterium tumefaciens Ti plasmids by quorum-sensing. TraM was overexpressed and purified from Escherichia coli. This protein binds to the transcriptional regulator TraR, abolishing its function. Size-exclusion chromatography and dynamic light scattering show that the recombinant protein has an apparent molecular […]
-
10/2002 - Analytical Chemistry
Determination of the stoichiometry of noncovalent complexes using reverse-phase high-performance liquid chromatography coupled with electrospray ion trap mass spectrometry
An electrospray mass spectrometry-based methodology has been developed to have a fast and sensitive method for protein-cofactor stoichiometry determination. As model systems, we used two proteins which require the presence of cofactors for activity: TraR, a member of the LuxR family of quorum-sensing transcriptional regulators, which requires an acyl-homoserine lactone molecule called Agrobacterium autoinducer (AAI) […]
-
09/2002 - EMBO J
The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA
The quorum sensing system allows bacteria to sense their cell density and initiate an altered pattern of gene expression after a sufficient quorum of cells has accumulated. In Agrobacterium tumefaciens, quorum sensing controls conjugal transfer of the tumour- inducing plasmid, responsible for plant crown gall disease. The core components of this system are the transcriptional […]