Vannini Group
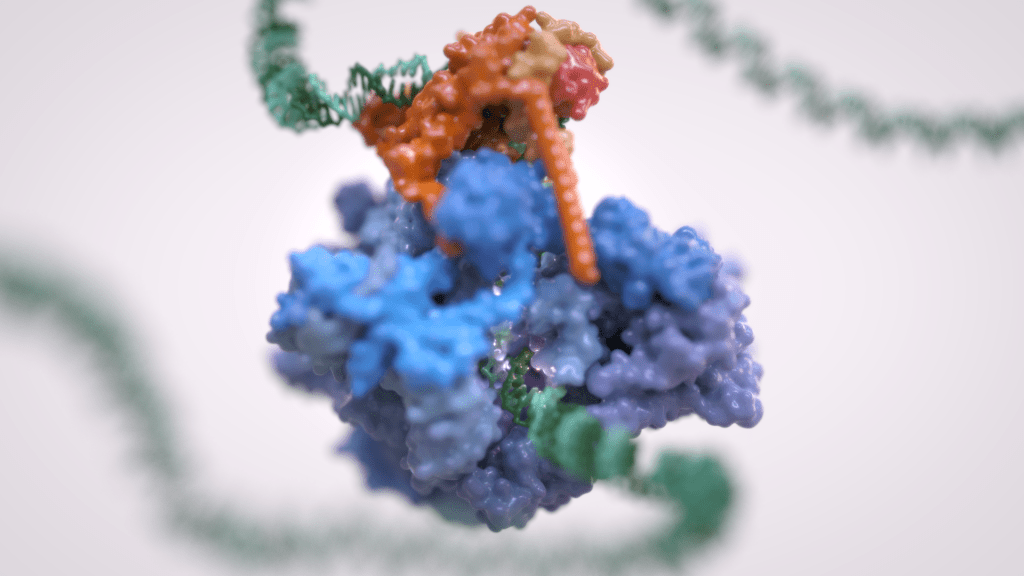
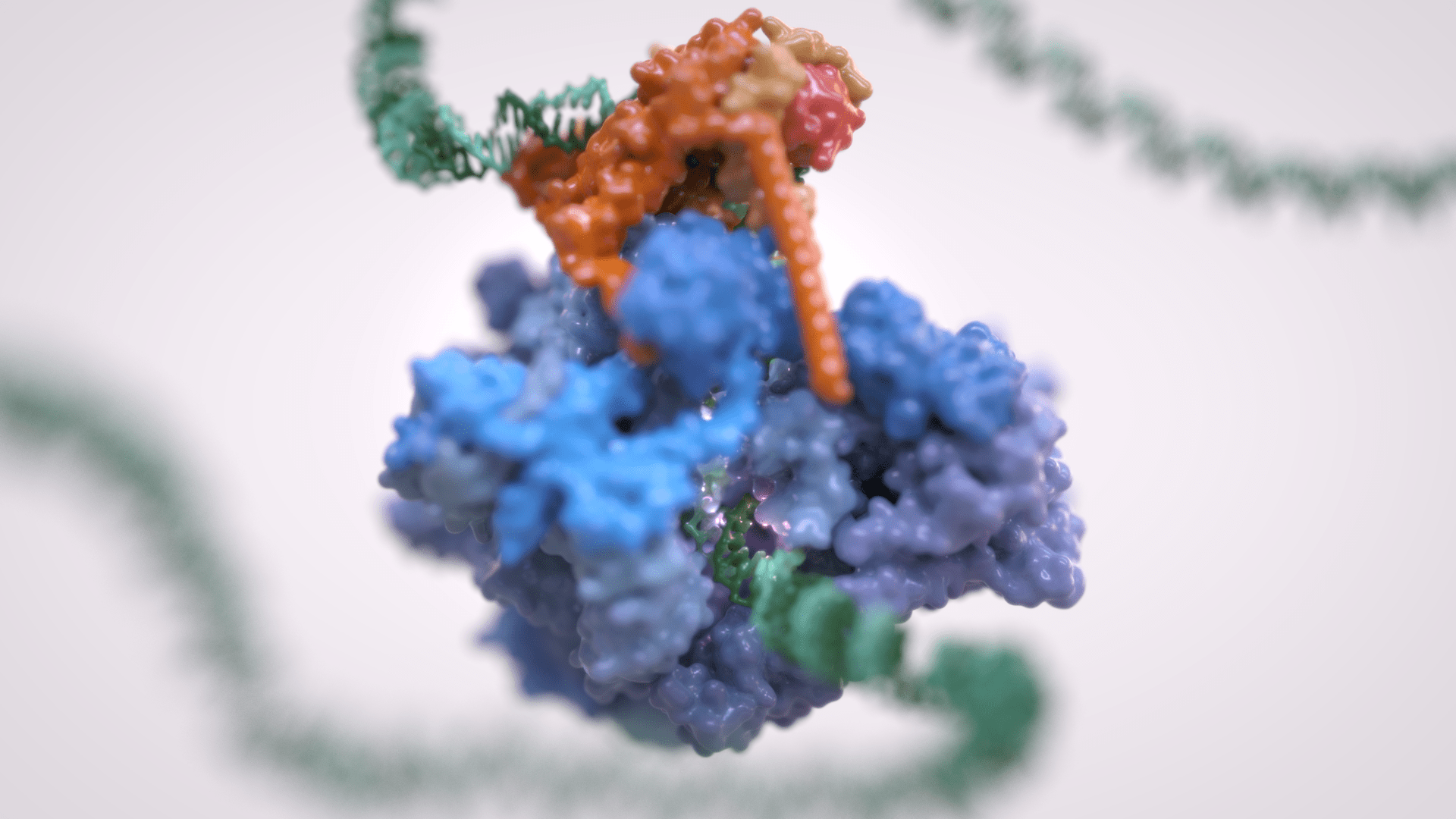
Gene transcription is the first step that controls the expression of the genetic information encoded in a genome and ultimately underlies cell differentiation and organism development. Eukaryotic gene transcription occurs in the context of highly structured and organised genomes and acts as a coordinator of numerous events co-occurring in the nucleus. Eukaryotic transcription relies on three different RNA polymerases: RNA polymerase I (Pol I) transcribes ribosomal RNA, RNA polymerase II (Pol II) synthesizes messenger RNAs and RNA polymerase III (Pol III) produces short and non-translated RNAs, including the entire pool of tRNAs, which are essential for cell growth.
For a long time, it was assumed that only Pol II was regulated whereas Pol I and Pol III did not require such control. However, it is now clear that RNA polymerase III transcription is tightly regulated and a determinant of organismal growth. Pol III deregulation is observed in many forms of cancer and Pol III genetic mutations cause severe neurodegenerative diseases.
Furthermore, Pol III and its associated factors play a paramount role into genome structure and organisation. These “extra-transcriptional roles” are carried out throughout interactions with other cellular components such as retroelement transposition machineries, Structural Maintenance of Chromosome (SMC) complexes and specific chromatin remodellers.
The Vannini Group employs an Integrative Structural Biology approach, combining cutting-edge cryo-EM analysis, x-ray diffraction data, cross-linking and native mass-spectrometry. We integrate the structural data with molecular and cellular biology techniques in order to obtain a comprehensive view of these fundamental processes and how their mis-regulation can lead to cancer and neurodegenerative diseases.
Group members
-
 Alessandro Vannini
Alessandro Vannini
Head of Structural Biology Research Centre -
 Alessandro Borsellini
Alessandro Borsellini
Postdoc -
 Giacomo Ettore Casale
Giacomo Ettore Casale
Postdoc -
 Valentina Cecatiello
Valentina Cecatiello
Senior Technician -
 Sebastian Chamera
Sebastian Chamera
Postdoc -
 Fabiola Iommazzo
Fabiola Iommazzo
PhD Student -
 Thomas Noé Perry
Thomas Noé Perry
Postdoc -
 Fabio Pessina
Fabio Pessina
Senior Technician -
 Mariavittoria Pizzinga
Mariavittoria Pizzinga
Postdoc -
 Ewan Ramsay
Ewan Ramsay
Senior Staff Scientist -
 Ankit Roy
Ankit Roy
PhD Student -
 Syed Zawar Shah
Syed Zawar Shah
PhD Student
Publications
-
12/2015 - Cell
Redox Signaling by the RNA Polymerase III TFIIB-Related Factor Brf2
TFIIB-related factor 2 (Brf2) is a member of the family of TFIIB-like core transcription factors. Brf2 recruits RNA polymerase (Pol) III to type III gene-external promoters, including the U6 spliceosomal RNA and selenocysteine tRNA genes. Found only in vertebrates, Brf2 has been linked to tumorigenesis but the underlying mechanisms remain elusive. We have solved crystal […]
-
09/2012 - Biochim Biophys Acta
A structural perspective on RNA polymerase I and RNA polymerase III transcription machineries
RNA polymerase I and III are responsible for the bulk of nuclear transcription in actively growing cells and their activity impacts the cellular biosynthetic capacity. As a consequence, RNA polymerase I and III deregulation has been directly linked to cancer development. The complexity of RNA polymerase I and III transcription apparatuses has hampered their structural […]
-
02/2012 - Molecular Cell
Conservation between the RNA polymerase I, II, and III transcription initiation machineries
Recent studies of the three eukaryotic transcription machineries revealed that all initiation complexes share a conserved core. This core consists of the RNA polymerase (I, II, or III), the TATA box-binding protein (TBP), and transcription factors TFIIB, TFIIE, and TFIIF (for Pol II) or proteins structurally and functionally related to parts of these factors (for […]
-
10/2010 - Cell
Molecular basis of RNA polymerase III transcription repression by Maf1
RNA polymerase III (Pol III) transcribes short RNAs required for cell growth. Under stress conditions, the conserved protein Maf1 rapidly represses Pol III transcription. We report the crystal structure of Maf1 and cryo-electron microscopic structures of Pol III, an active Pol III-DNA-RNA complex, and a repressive Pol III-Maf1 complex. Binding of DNA and RNA causes […]
-
10/2010 - Nucleic Acids Res
The archaeo-eukaryotic primase of plasmid pRN1 requires a helix bundle domain for faithful primer synthesis
The plasmid pRN1 encodes for a multifunctional replication protein with primase, DNA polymerase and helicase activity. The minimal region required for primase activity encompasses amino-acid residues 40-370. While the N-terminal part of that minimal region (residues 47-247) folds into the prim/pol domain and bears the active site, the structure and function of the C-terminal part […]